Gene Page: IGF1R
Summary ?
| GeneID | 3480 |
| Symbol | IGF1R |
| Synonyms | CD221|IGFIR|IGFR|JTK13 |
| Description | insulin like growth factor 1 receptor |
| Reference | MIM:147370|HGNC:HGNC:5465|Ensembl:ENSG00000140443|HPRD:00932|Vega:OTTHUMG00000149851 |
| Gene type | protein-coding |
| Map location | 15q26.3 |
| Pascal p-value | 0.125 |
| Sherlock p-value | 0.037 |
| Fetal beta | 2.226 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0073 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10507754 | 15 | 99504358 | IGF1R | 2.957E-4 | 0.319 | 0.039 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4965530 | chr15 | 98720317 | IGF1R | 3480 | 0.2 | cis |
Section II. Transcriptome annotation
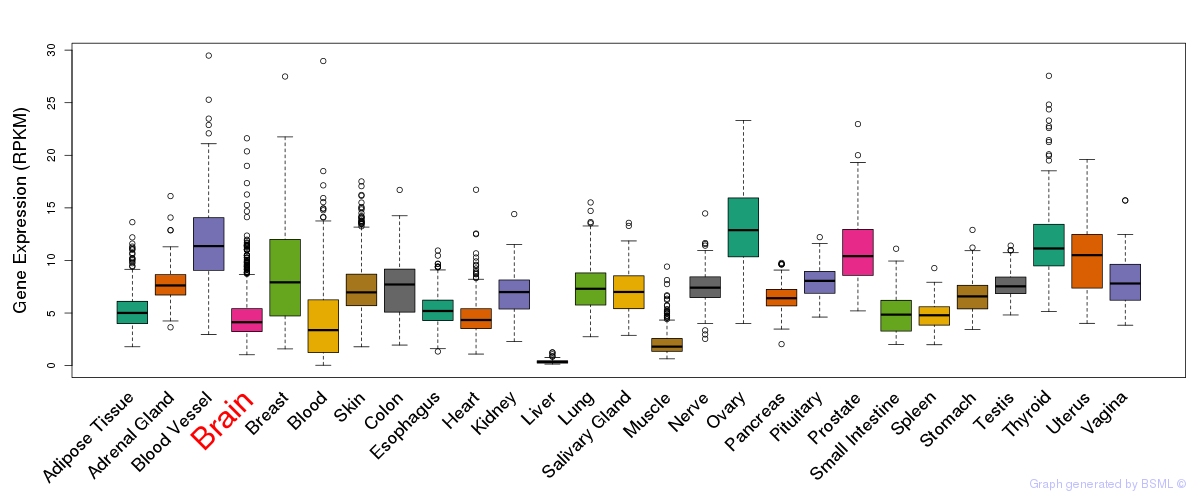
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005010 | insulin-like growth factor receptor activity | IDA | 7679099 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005158 | insulin receptor binding | IDA | 8452530 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 11448933 | |
| GO:0031994 | insulin-like growth factor I binding | IPI | 8452530 | |
| GO:0043548 | phosphoinositide 3-kinase binding | IPI | 7541045 | |
| GO:0043559 | insulin binding | IPI | 8452530 | |
| GO:0043560 | insulin receptor substrate binding | IPI | 7541045 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0030238 | male sex determination | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0007165 | signal transduction | TAS | 3003744 | |
| GO:0008284 | positive regulation of cell proliferation | TAS | 10749889 | |
| GO:0008286 | insulin receptor signaling pathway | TAS | 10829031 | |
| GO:0048015 | phosphoinositide-mediated signaling | IDA | 7692086 | |
| GO:0048009 | insulin-like growth factor receptor signaling pathway | IDA | 7679099 | |
| GO:0006955 | immune response | IMP | 16886151 | |
| GO:0006916 | anti-apoptosis | TAS | 8710868 | |
| GO:0014065 | phosphoinositide 3-kinase cascade | IC | 7541045 | |
| GO:0051262 | protein tetramerization | IDA | 1846292 | |
| GO:0030335 | positive regulation of cell migration | IMP | 12138094 | |
| GO:0045740 | positive regulation of DNA replication | IMP | 12138094 | |
| GO:0046777 | protein amino acid autophosphorylation | IDA | 1846292 |7679099 |11162456 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005792 | microsome | IDA | 8452530 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | IC | 7679099 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGEF12 | DKFZp686O2372 | KIAA0382 | LARG | PRO2792 | Rho guanine nucleotide exchange factor (GEF) 12 | - | HPRD,BioGRID | 11724822 |
| CSK | MGC117393 | c-src tyrosine kinase | CSK interacts with IGF-IR. | BIND | 10026153 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 10026153 |
| DOK5 | C20orf180 | MGC16926 | docking protein 5 | - | HPRD | 12730241 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | Affinity Capture-Western | BioGRID | 17047074 |
| EHD1 | FLJ42622 | FLJ44618 | H-PAST | HPAST1 | PAST | PAST1 | EH-domain containing 1 | - | HPRD,BioGRID | 11423532 |
| EHD1 | FLJ42622 | FLJ44618 | H-PAST | HPAST1 | PAST | PAST1 | EH-domain containing 1 | EHD1 interacts with IGF-1R. | BIND | 11423532 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Affinity Capture-Western | BioGRID | 16113100 |
| GIGYF1 | GYF1 | PERQ1 | GRB10 interacting GYF protein 1 | - | HPRD | 12771153 |
| GNAI1 | Gi | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | - | HPRD,BioGRID | 10644671 |
| GNAI2 | GIP | GNAI2B | H_LUCA15.1 | H_LUCA16.1 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 | - | HPRD | 11120746 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | IGF-IR interacts with RACK1. | BIND | 11884618 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 11884618 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD | 8621530 |8764099 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | Grb10 interacts with IGFIR. | BIND | 9506989 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 8764099 |8776723 |9506989 |12697834 |
| IGF1 | IGFI | insulin-like growth factor 1 (somatomedin C) | IGF-I interacts with IGF-IR. This interaction was modeled on a demonstrated interaction between IGF-I from an unspecified species and human IGF-IR. | BIND | 15604363 |
| IGF1 | IGFI | insulin-like growth factor 1 (somatomedin C) | - | HPRD,BioGRID | 1852007 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD | 12138114 |
| IGF2 | C11orf43 | FLJ22066 | FLJ44734 | INSIGF | pp9974 | insulin-like growth factor 2 (somatomedin A) | - | HPRD,BioGRID | 9972281 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | - | HPRD,BioGRID | 9389554 |
| INS | ILPR | IRDN | insulin | Insulin interacts with IGF-IR. This interaction was modeled on a demonstrated interaction between Insulin from an unspecified species and human IGF-IR. | BIND | 15604363 |
| INS | ILPR | IRDN | insulin | - | HPRD | 1851182 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | IGFIR interacts with IRS-1. | BIND | 7541045 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | IRS-1 interacts with IGF-IR. This interaction was modeled on a demonstrated interaction between rat IRS-1 and human IGF-IR. | BIND | 10026153 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 7559507 |
| IRS2 | - | insulin receptor substrate 2 | - | HPRD | 9852124 |
| IRS2 | - | insulin receptor substrate 2 | Insulin Receptor Substrate-2 Binds to the Insulin-like Growth Factor Receptor Beta subunit. This interaction is modeled on demonstrated interaction between human IGF-IR and murine IRS-2 | BIND | 8626379 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | beta-1 integrin interacts with IGF-IR. | BIND | 11884618 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | Phosphorylated IGF-1R-beta interacts with JAK-1. | BIND | 9492017 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD,BioGRID | 9492017 |
| NAMPT | 1110035O14Rik | DKFZp666B131 | MGC117256 | PBEF | PBEF1 | VF | VISFATIN | nicotinamide phosphoribosyltransferase | PBEF interacts with IGF-IR. This interaction was modeled on a demonstrated interaction between PBEF from an unspecified species and human IGF-IR. | BIND | 15604363 |
| NEDD4 | KIAA0093 | MGC176705 | NEDD4-1 | RPF1 | neural precursor cell expressed, developmentally down-regulated 4 | - | HPRD,BioGRID | 12697834 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 8697095 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | IGFIR interacts with p85. This interaction was modeled on a demonstrated interaction between rat proteins. | BIND | 7541045 |
| PIK3R3 | DKFZp686P05226 | FLJ41892 | p55 | p55-GAMMA | phosphoinositide-3-kinase, regulatory subunit 3 (gamma) | - | HPRD,BioGRID | 9415396 |9524259 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | PKC-delta interacts with IGF-IR. | BIND | 11884618 |
| PRKD1 | PKC-MU | PKCM | PKD | PRKCM | protein kinase D1 | PKC-mu interacts with IGF-IR. | BIND | 11884618 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western | BioGRID | 10082579 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD,BioGRID | 8999839 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD,BioGRID | 7642582 |
| PXN | FLJ16691 | paxillin | Affinity Capture-Western | BioGRID | 10082579 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD,BioGRID | 7642582 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | IGFIR interacts with SHC. | BIND | 7541045 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 8776723 |
| SNAP29 | CEDNIK | FLJ21051 | SNAP-29 | synaptosomal-associated protein, 29kDa | - | HPRD,BioGRID | 11423532 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | - | HPRD,BioGRID | 9727029 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | Insulin-like Growth Factor-I Receptor interacts with SOCS1 | BIND | 9727029 |
| SOCS2 | CIS2 | Cish2 | SOCS-2 | SSI-2 | SSI2 | STATI2 | suppressor of cytokine signaling 2 | Interaction of Human Suppressor of Cytokine Signaling (SOCS)-2 with the Insulin-like Growth Factor-I Receptor | BIND | 9727029 |
| SOCS2 | CIS2 | Cish2 | SOCS-2 | SSI-2 | SSI2 | STATI2 | suppressor of cytokine signaling 2 | - | HPRD,BioGRID | 9727029 |
| SOCS3 | ATOD4 | CIS3 | Cish3 | MGC71791 | SOCS-3 | SSI-3 | SSI3 | suppressor of cytokine signaling 3 | Suppressor of cytokine signaling (SOCS)-3 protein interacts with the insulin-like growth factor I receptor. | BIND | 11071852 |
| SOCS3 | ATOD4 | CIS3 | Cish3 | MGC71791 | SOCS-3 | SSI-3 | SSI3 | suppressor of cytokine signaling 3 | in vitro in vivo | BioGRID | 11071852 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | - | HPRD,BioGRID | 11094073 |
| WISP2 | CCN5 | CT58 | CTGF-L | WNT1 inducible signaling pathway protein 2 | - | HPRD | 10358067 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD,BioGRID | 9581554 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | IGFIR interacts with 14-3-3-beta isoform. This interaction was modelled on a demonstrated interaction between human IGFIR and mouse 14-3-3-beta. | BIND | 9581554 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD,BioGRID | 9111084 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | IGFIR interacts with 14-3-3-epsilon isoform. This interaction is modeled on a demonstrated interaction between human IGFIR and mouse 14-3-3-epsilon isoform | BIND | 9111084 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD | 9581554 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | IGFIR interacts with 14-3-3-zeta isoform. | BIND | 9581554 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG PROGESTERONE MEDIATED OOCYTE MATURATION | 86 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1 PATHWAY | 21 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1R PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BAD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1MTOR PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TEL PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LONGEVITY PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| PID ER NONGENOMIC PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID IGF1 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| WATANABE COLON CANCER MSI VS MSS DN | 81 | 42 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| BERENJENO ROCK SIGNALING NOT VIA RHOA DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BARIS THYROID CANCER DN | 59 | 45 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| LI CISPLATIN RESISTANCE UP | 28 | 20 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM UP | 81 | 57 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE IMMORTALIZED DN | 31 | 26 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 15Q26 AMPLICON | 22 | 19 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE DN | 52 | 37 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS UP | 81 | 54 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| GENTILE UV LOW DOSE DN | 67 | 46 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4 | 94 | 69 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GAMMA RADIATION RESISTANCE | 20 | 14 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCERS KINOME BLUE | 21 | 16 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL | 61 | 47 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 1 | 68 | 50 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR UP | 61 | 44 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS UP | 61 | 41 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS UP | 68 | 49 | All SZGR 2.0 genes in this pathway |
| FARDIN HYPOXIA 11 | 32 | 29 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 2619 | 2626 | 1A,m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU | ||||
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-122 | 1347 | 1353 | 1A | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-143 | 2664 | 2670 | 1A | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-145 | 3804 | 3810 | m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-15/16/195/424/497 | 3502 | 3508 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-153 | 188 | 194 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-182 | 586 | 593 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA | ||||
| miR-194 | 6813 | 6819 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-214 | 1870 | 1876 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-217 | 152 | 158 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-223 | 224 | 231 | 1A,m8 | hsa-miR-223 | UGUCAGUUUGUCAAAUACCCC |
| miR-30-5p | 5798 | 5804 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-320 | 196 | 202 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA | ||||
| miR-326 | 5069 | 5075 | 1A | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-363 | 5748 | 5754 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-376 | 1307 | 1314 | 1A,m8 | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-376c | 1162 | 1168 | m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-378* | 5589 | 5595 | m8 | hsa-miR-422b | CUGGACUUGGAGUCAGAAGGCC |
| hsa-miR-422a | CUGGACUUAGGGUCAGAAGGCC | ||||
| miR-381 | 6905 | 6911 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-409-5p | 2706 | 2712 | m8 | hsa-miR-409-5p | AGGUUACCCGAGCAACUUUGCA |
| miR-410 | 6863 | 6869 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-421 | 4195 | 4201 | 1A | hsa-miR-421 | GGCCUCAUUAAAUGUUUGUUG |
| miR-448 | 187 | 194 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-455 | 3782 | 3788 | 1A | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-494 | 5648 | 5655 | 1A,m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-495 | 6811 | 6817 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-503 | 1269 | 1275 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 2211 | 2217 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
| miR-96 | 5822 | 5828 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
| hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC | ||||
| miR-99/100 | 5602 | 5609 | 1A,m8 | hsa-miR-99abrain | AACCCGUAGAUCCGAUCUUGUG |
| hsa-miR-100brain | AACCCGUAGAUCCGAACUUGUG | ||||
| hsa-miR-99bbrain | CACCCGUAGAACCGACCUUGCG | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.