Gene Page: MYC
Summary ?
| GeneID | 4609 |
| Symbol | MYC |
| Synonyms | MRTL|MYCC|bHLHe39|c-Myc |
| Description | v-myc avian myelocytomatosis viral oncogene homolog |
| Reference | MIM:190080|HGNC:HGNC:7553|Ensembl:ENSG00000136997|HPRD:01818|Vega:OTTHUMG00000128475 |
| Gene type | protein-coding |
| Map location | 8q24.21 |
| Pascal p-value | 3.014E-4 |
| Fetal beta | 2.211 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.408 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0359 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16100135 | 8 | 128750586 | MYC | 3.48E-9 | -0.02 | 2.27E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10094528 | chr8 | 9740328 | MYC | 4609 | 0.08 | trans | ||
| rs7107744 | chr11 | 128268778 | MYC | 4609 | 0.1 | trans | ||
| rs837543 | chr16 | 55500411 | MYC | 4609 | 0.06 | trans |
Section II. Transcriptome annotation
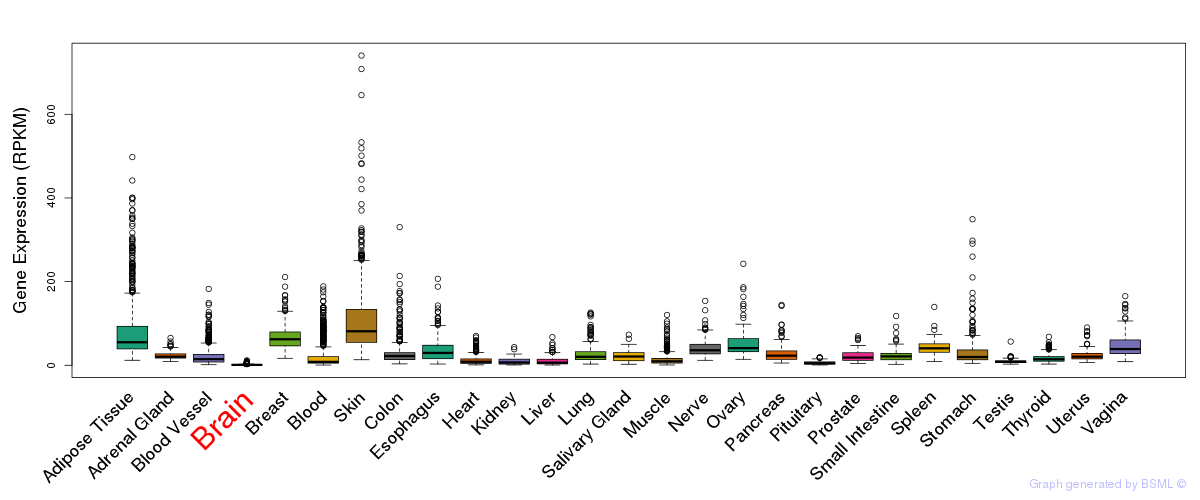
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TSKU | 0.69 | 0.71 |
| C3orf54 | 0.64 | 0.71 |
| TTLL12 | 0.64 | 0.62 |
| NEUROD2 | 0.64 | 0.59 |
| TP53I11 | 0.63 | 0.67 |
| PTPN2 | 0.63 | 0.64 |
| NKAIN1 | 0.63 | 0.68 |
| NXPH3 | 0.63 | 0.47 |
| DBN1 | 0.62 | 0.66 |
| IMPDH2 | 0.62 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FBXO2 | -0.47 | -0.68 |
| HLA-F | -0.46 | -0.66 |
| AIFM3 | -0.46 | -0.64 |
| C5orf53 | -0.45 | -0.63 |
| ARHGAP22 | -0.45 | -0.65 |
| LDHD | -0.45 | -0.63 |
| ALDOC | -0.44 | -0.62 |
| CLU | -0.44 | -0.63 |
| HEPN1 | -0.44 | -0.62 |
| CSRP1 | -0.44 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | NAS | 2834731 | |
| GO:0003700 | transcription factor activity | TAS | 9924025 | |
| GO:0005515 | protein binding | IPI | 9308237 |9708738 |10597290 |12706874 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001836 | release of cytochrome c from mitochondria | IEA | - | |
| GO:0001783 | B cell apoptosis | IEA | - | |
| GO:0006309 | DNA fragmentation during apoptosis | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | NAS | 2834731 | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 9924025 | |
| GO:0007050 | cell cycle arrest | TAS | 10962037 | |
| GO:0008284 | positive regulation of cell proliferation | IDA | 15994933 | |
| GO:0008629 | induction of apoptosis by intracellular signals | IEA | - | |
| GO:0008633 | activation of pro-apoptotic gene products | IEA | - | |
| GO:0008634 | negative regulation of survival gene product expression | IEA | - | |
| GO:0006919 | caspase activation | IEA | - | |
| GO:0006879 | cellular iron ion homeostasis | TAS | 9924025 | |
| GO:0009314 | response to radiation | IEA | - | |
| GO:0042981 | regulation of apoptosis | IEA | - | |
| GO:0016485 | protein processing | IEA | - | |
| GO:0042474 | middle ear morphogenesis | IEA | - | |
| GO:0043473 | pigmentation | IEA | - | |
| GO:0043279 | response to alkaloid | IEA | - | |
| GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | IEA | - | |
| GO:0048705 | skeletal system morphogenesis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005819 | spindle | IEA | - | |
| GO:0005634 | nucleus | IDA | 15994933 | |
| GO:0005634 | nucleus | NAS | 2834731 | |
| GO:0016604 | nuclear body | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | Co-purification | BioGRID | 11839798 |
| ACTL6A | ACTL6 | Arp4 | BAF53A | INO80K | MGC5382 | actin-like 6A | - | HPRD,BioGRID | 11839798 |
| ARHGEF2 | DKFZp547L106 | DKFZp547P1516 | GEF | GEF-H1 | GEFH1 | KIAA0651 | LFP40 | P40 | rho/rac guanine nucleotide exchange factor (GEF) 2 | Affinity Capture-MS | BioGRID | 17353931 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | Affinity Capture-Western Co-localization | BioGRID | 15210690 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 9788437 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 association with c-Myc, a proto-oncogene that is implicated in tumorigenesis, embryonic development and apoptosis, inhibits its transcriptional and transforming activity in cells. | BIND | 9788437 |
| CCNT1 | CCNT | CYCT1 | cyclin T1 | - | HPRD,BioGRID | 12944920 |
| CD6 | FLJ44171 | TP120 | CD6 molecule | Affinity Capture-Western | BioGRID | 10899308 |
| CDC6 | CDC18L | HsCDC18 | HsCDC6 | cell division cycle 6 homolog (S. cerevisiae) | - | HPRD,BioGRID | 10899308 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | MYC (c-Myc) interacts with CDK4 promoter. | BIND | 12857727 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | Affinity Capture-Western | BioGRID | 12944920 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Myc interacts with CDKN1A promoter. | BIND | 15780936 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | c-Myc binds the proximal region of the p21Cip1 promoter. | BIND | 15084259 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | c-Myc interacts with p21WAF1 promoter. | BIND | 15856024 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | p14ARF interacts with c-Myc. | BIND | 15361884 |
| CEBPA | C/EBP-alpha | CEBP | CCAAT/enhancer binding protein (C/EBP), alpha | - | HPRD,BioGRID | 12873812 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 12873812 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | IKK-alpha interacts with c-myc promoter. | BIND | 15808510 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | CK2 interacts with and phosphorylates c-Myc. This interaction was modelled on a demonstrated interaction between mouse CK2 and human c-Myc. | BIND | 12149649 |
| DDB2 | DDBB | FLJ34321 | UV-DDB2 | damage-specific DNA binding protein 2, 48kDa | c-Myc interacts with DDB2 promoter. | BIND | 15856024 |
| DDX24 | - | DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 | Affinity Capture-MS | BioGRID | 17353931 |
| DNMT3A | DNMT3A2 | M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha | Myc interacts with Dmnt3a. | BIND | 15616584 |
| DNMT3A | DNMT3A2 | M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha | - | HPRD,BioGRID | 15616584 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | E2F-1 interacts with the MYC gene P2 promoter region. | BIND | 7892279 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | E2F-4 interacts with the MYC gene P2 promoter region. | BIND | 7892279 |
| E2F5 | E2F-5 | E2F transcription factor 5, p130-binding | E2F-5 interacts with the MYC gene P2 promoter region. | BIND | 7892279 |
| EFTUD2 | DKFZp686E24196 | FLJ44695 | KIAA0031 | Snrp116 | Snu114 | U5-116KD | elongation factor Tu GTP binding domain containing 2 | Affinity Capture-MS | BioGRID | 17353931 |
| EP400 | CAGH32 | DKFZP434I225 | FLJ42018 | FLJ45115 | P400 | TNRC12 | E1A binding protein p400 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11509179 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ER-alpha interacts with c-myc promoter. | BIND | 15808510 |
| EXOC4 | MGC27170 | REC8 | SEC8 | SEC8L1 | Sec8p | exocyst complex component 4 | Affinity Capture-MS | BioGRID | 17353931 |
| FANCI | FLJ10719 | KIAA1794 | Fanconi anemia, complementation group I | Affinity Capture-MS | BioGRID | 17353931 |
| FBXW7 | AGO | CDC4 | DKFZp686F23254 | FBW6 | FBW7 | FBX30 | FBXO30 | FBXW6 | FLJ16457 | SEL-10 | SEL10 | F-box and WD repeat domain containing 7 | Fbw7-alpha interacts with c-Myc. | BIND | 16023596 |
| FBXW7 | AGO | CDC4 | DKFZp686F23254 | FBW6 | FBW7 | FBX30 | FBXO30 | FBXW6 | FLJ16457 | SEL-10 | SEL10 | F-box and WD repeat domain containing 7 | Fbw7-beta interacts with c-Myc. | BIND | 16023596 |
| FUBP1 | FBP | FUBP | far upstream element (FUSE) binding protein 1 | FBP interacts with FUSE. | BIND | 15502847 |
| GCN1L1 | GCN1 | GCN1L | KIAA0219 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | Affinity Capture-MS | BioGRID | 17353931 |
| GTF2B | TF2B | TFIIB | general transcription factor IIB | Reconstituted Complex | BioGRID | 12660246 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | - | HPRD,BioGRID | 8755740 |
| GTF2I | BAP-135 | BAP135 | BTKAP1 | DIWS | FLJ38776 | FLJ56355 | IB291 | SPIN | TFII-I | WBS | WBSCR6 | general transcription factor II, i | - | HPRD,BioGRID | 8377829 |
| GTF3C3 | TFIIIC102 | TFIIICgamma | TFiiiC2-102 | general transcription factor IIIC, polypeptide 3, 102kDa | Affinity Capture-MS | BioGRID | 17353931 |
| HEATR1 | BAP28 | FLJ10359 | MGC72083 | HEAT repeat containing 1 | Affinity Capture-MS | BioGRID | 17353931 |
| HNRNPC | C1 | C2 | HNRNP | HNRPC | MGC104306 | MGC105117 | MGC117353 | MGC131677 | SNRPC | heterogeneous nuclear ribonucleoprotein C (C1/C2) | Affinity Capture-MS | BioGRID | 17353931 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 12644583 |
| KAT2A | GCN5 | GCN5L2 | MGC102791 | PCAF-b | hGCN5 | K(lysine) acetyltransferase 2A | c-Myc interacts with GNC5L. | BIND | 12660246 |
| KAT2A | GCN5 | GCN5L2 | MGC102791 | PCAF-b | hGCN5 | K(lysine) acetyltransferase 2A | Affinity Capture-Western | BioGRID | 10611234 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 12660246 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | - | HPRD,BioGRID | 12776177 |
| KIAA0859 | 5630401D24Rik | CGI-01 | FLJ10310 | KIAA0859 | Affinity Capture-MS | BioGRID | 17353931 |
| KIAA1967 | DBC-1 | DBC1 | KIAA1967 | Affinity Capture-MS | BioGRID | 17353931 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | - | HPRD | 7957875 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Biochemical Activity Reconstituted Complex | BioGRID | 7957875 |9207092 |15210690 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | Reconstituted Complex | BioGRID | 15210690 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | ERK1 interacts with and phosphorylates c-Myc. | BIND | 9155018 |
| MAPK7 | BMK1 | ERK4 | ERK5 | PRKM7 | mitogen-activated protein kinase 7 | - | HPRD | 9461566 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | - | HPRD,BioGRID | 10551811 |
| MAX | MGC10775 | MGC11225 | MGC18164 | MGC34679 | MGC36767 | bHLHd4 | orf1 | MYC associated factor X | Myc interacts with Max. This interaction was modeled on a demonstrated interaction between human Myc and Max from an unspecified source. | BIND | 12584560 |
| MAX | MGC10775 | MGC11225 | MGC18164 | MGC34679 | MGC36767 | bHLHd4 | orf1 | MYC associated factor X | Affinity Capture-MS Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 2006410 |9528857 |9708738 |10229200 |10319872 |10593926 |10611234 |12391307 |12584560 |17353931 |
| MAX | MGC10775 | MGC11225 | MGC18164 | MGC34679 | MGC36767 | bHLHd4 | orf1 | MYC associated factor X | - | HPRD | 2006410 |10229200 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | Affinity Capture-MS | BioGRID | 17353931 |
| MED12 | CAGH45 | HOPA | KIAA0192 | OPA1 | TNRC11 | TRAP230 | mediator complex subunit 12 | Reconstituted Complex | BioGRID | 12660246 |
| MED14 | CRSP150 | CRSP2 | CSRP | CXorf4 | DRIP150 | EXLM1 | MGC104513 | RGR1 | TRAP170 | mediator complex subunit 14 | Reconstituted Complex | BioGRID | 12660246 |
| MINA | DKFZp762O1912 | FLJ14393 | MDIG | MINA53 | NO52 | MYC induced nuclear antigen | - | HPRD | 12091391 |
| MIRHG1 | C13orf25 | FLJ14178 | MGC126270 | MIHG1 | MIRH1 | NCRNA00048 | microRNA host gene 1 (non-protein coding) | Myc interacts with the mir-17 cluster locus | BIND | 15944709 |
| MKLN1 | FLJ11162 | TWA2 | muskelin 1, intracellular mediator containing kelch motifs | Affinity Capture-MS | BioGRID | 17353931 |
| MLH1 | COCA2 | FCC2 | HNPCC | HNPCC2 | MGC5172 | hMLH1 | mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) | - | HPRD,BioGRID | 12584560 |
| MLH1 | COCA2 | FCC2 | HNPCC | HNPCC2 | MGC5172 | hMLH1 | mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) | MLH1 interacts with Myc. | BIND | 12584560 |
| MSH2 | COCA1 | FCC1 | HNPCC | HNPCC1 | LCFS2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | Affinity Capture-Western | BioGRID | 12584560 |
| MSH6 | GTBP | HNPCC5 | HSAP | mutS homolog 6 (E. coli) | Affinity Capture-MS | BioGRID | 17353931 |
| MXD1 | MAD | MAD1 | MGC104659 | MAX dimerization protein 1 | - | HPRD,BioGRID | 11602341 |
| MYBBP1A | FLJ37886 | P160 | PAP2 | MYB binding protein (P160) 1a | Affinity Capture-MS | BioGRID | 17353931 |
| MYCBP | AMY-1 | c-myc binding protein | - | HPRD,BioGRID | 9797456 |
| MYCBP2 | DKFZp686M08244 | FLJ10106 | FLJ13826 | FLJ21597 | FLJ21646 | KIAA0916 | PAM | MYC binding protein 2 | Myc interacts with Pam. | BIND | 9689053 |
| MYCBP2 | DKFZp686M08244 | FLJ10106 | FLJ13826 | FLJ21597 | FLJ21646 | KIAA0916 | PAM | MYC binding protein 2 | - | HPRD,BioGRID | 9689053 |
| MYO1B | myr1 | myosin IB | Affinity Capture-MS | BioGRID | 17353931 |
| NCL | C23 | FLJ45706 | nucleolin | Ncl (nucleolin) interacts with Myc (c-Myc). | BIND | 15674325 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | AIB1 interacts with c-myc promoter. | BIND | 15808510 |
| NFYB | CBF-A | CBF-B | HAP3 | NF-YB | nuclear transcription factor Y, beta | - | HPRD,BioGRID | 11282029 |
| NFYB | CBF-A | CBF-B | HAP3 | NF-YB | nuclear transcription factor Y, beta | Myc interacts with NF-YB. This interaction was modeled on a demonstrated interaction between human Myc and mouse NF-YB | BIND | 11282029 |
| NFYC | CBF-C | CBFC | DKFZp667G242 | FLJ45775 | H1TF2A | HAP5 | HSM | NF-YC | nuclear transcription factor Y, gamma | Myc interacts with NF-YC. | BIND | 11282029 |
| NFYC | CBF-C | CBFC | DKFZp667G242 | FLJ45775 | H1TF2A | HAP5 | HSM | NF-YC | nuclear transcription factor Y, gamma | - | HPRD,BioGRID | 10446203 |
| NMI | - | N-myc (and STAT) interactor | - | HPRD | 8668343 |
| NMI | - | N-myc (and STAT) interactor | Affinity Capture-Western Reconstituted Complex | BioGRID | 11916966 |
| NMI | - | N-myc (and STAT) interactor | Myc interacts with Nmi | BIND | 11916966 |
| NPM1 | B23 | MGC104254 | NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | Myc interacts with NPM1 (B23) | BIND | 15944709 |
| NUP205 | C7orf14 | nucleoporin 205kDa | Affinity Capture-MS | BioGRID | 17353931 |
| NUP93 | KIAA0095 | MGC21106 | nucleoporin 93kDa | Affinity Capture-MS | BioGRID | 17353931 |
| ORC2L | ORC2 | origin recognition complex, subunit 2-like (yeast) | Ocr2 interacts with C-Myc origin. | BIND | 15910003 |
| PARP10 | FLJ14464 | poly (ADP-ribose) polymerase family, member 10 | PARP10 interacts with Myc (c-Myc) | BIND | 15674325 |
| PDS5A | DKFZp686B19246 | FLJ41012 | KIAA0648 | MGC131948 | MGC161503 | PIG54 | SCC-112 | PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| PFDN5 | MGC5329 | MGC71907 | MM-1 | MM1 | PFD5 | prefoldin subunit 5 | Reconstituted Complex Two-hybrid | BioGRID | 9792694 |11567024 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Affinity Capture-Western | BioGRID | 11283613 |
| PMAIP1 | APR | NOXA | phorbol-12-myristate-13-acetate-induced protein 1 | c-Myc interacts with NOXA promoter. | BIND | 15856024 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | Myc interacts with PML. This interaction was modelled on a demonstrated interaction between mouse Myc and human PML. | BIND | 15735755 |
| POLE | DKFZp434F222 | FLJ21434 | POLE1 | polymerase (DNA directed), epsilon | Affinity Capture-MS | BioGRID | 17353931 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Pol II interacts with c-myc promoter. | BIND | 15808510 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Reconstituted Complex | BioGRID | 12660246 |
| PRPF8 | HPRP8 | PRP8 | PRPC8 | RP13 | PRP8 pre-mRNA processing factor 8 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 9315742 |
| RAGE | MOK | RAGE1 | renal tumor antigen | - | HPRD,BioGRID | 10421840 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 7838535 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | - | HPRD,BioGRID | 8076603 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 12027803 |
| RIBIN | - | rRNA promoter binding protein | c-Myc interacts with rDNA. | BIND | 15723054 |
| RRM2B | DKFZp686M05248 | MGC102856 | MGC42116 | p53R2 | ribonucleotide reductase M2 B (TP53 inducible) | c-Myc interacts with p53R2 promoter. | BIND | 15856024 |
| RUVBL1 | ECP54 | INO80H | NMP238 | PONTIN | Pontin52 | RVB1 | TIH1 | TIP49 | TIP49A | RuvB-like 1 (E. coli) | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 11509179 |11839798 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | Co-purification | BioGRID | 11839798 |
| SAP130 | FLJ12761 | Sin3A-associated protein, 130kDa | Co-purification Reconstituted Complex | BioGRID | 12660246 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 11804592 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 11804592 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11839798 |17353931 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | - | HPRD,BioGRID | 10319872 |
| SNRNP200 | ASCC3L1 | BRR2 | HELIC2 | U5-200KD | small nuclear ribonucleoprotein 200kDa (U5) | Affinity Capture-MS | BioGRID | 17353931 |
| SP1 | - | Sp1 transcription factor | Myc interacts with Sp1. This interaction was modeled on a demonstrated interaction between mouse Myc and human Sp-1. | BIND | 11274368 |
| SP1 | - | Sp1 transcription factor | Sp1 interacts with Myc. | BIND | 15780936 |
| SP1 | - | Sp1 transcription factor | - | HPRD,BioGRID | 11274368 |
| SPAG9 | FLJ13450 | FLJ14006 | FLJ26141 | FLJ34602 | HLC4 | JLP | KIAA0516 | MGC117291 | MGC14967 | MGC74461 | PHET | PIG6 | sperm associated antigen 9 | - | HPRD | 12391307 |
| SUPT3H | SPT3 | SPT3L | suppressor of Ty 3 homolog (S. cerevisiae) | - | HPRD,BioGRID | 12660246 |
| TADA2L | ADA2 | ADA2A | FLJ12705 | KL04P | hADA2 | transcriptional adaptor 2 (ADA2 homolog, yeast)-like | Co-purification Reconstituted Complex | BioGRID | 12660246 |
| TADA3L | ADA3 | FLJ20221 | FLJ21329 | hADA3 | transcriptional adaptor 3 (NGG1 homolog, yeast)-like | Co-purification | BioGRID | 12660246 |
| TAF10 | TAF2A | TAF2H | TAFII30 | TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa | Co-purification | BioGRID | 12660246 |
| TAF12 | TAF2J | TAFII20 | TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa | Co-purification | BioGRID | 12660246 |
| TAF1A | MGC:17061 | RAFI48 | SL1 | TAFI48 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa | TAFI48 interacts with c-Myc. | BIND | 15723054 |
| TAF1B | MGC:9349 | RAF1B | RAFI63 | SL1 | TAFI63 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa | TAFI63 interacts with c-Myc. | BIND | 15723054 |
| TAF1C | MGC:39976 | SL1 | TAFI110 | TAFI95 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa | TAFI110 interacts with c-Myc. | BIND | 15723054 |
| TAF5 | TAF2D | TAFII100 | TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa | Reconstituted Complex | BioGRID | 12660246 |
| TAF5L | PAF65B | TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa | Co-purification | BioGRID | 12660246 |
| TAF6L | FLJ11136 | MGC4288 | PAF65A | TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa | Co-purification | BioGRID | 12660246 |
| TAF9 | AD-004 | AK6 | CGI-137 | CINAP | CIP | MGC1603 | MGC3647 | MGC5067 | MGC:1603 | MGC:3647 | MGC:5067 | TAF2G | TAFII31 | TAFII32 | TAFIID32 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 12660246 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | Reconstituted Complex | BioGRID | 8755740 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | TBP interacts with c-Myc. | BIND | 15723054 |
| TFAP2A | AP-2 | AP-2alpha | AP2TF | BOFS | TFAP2 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | Affinity Capture-Western Reconstituted Complex | BioGRID | 7729426 |
| TFAP2B | AP-2B | AP2-B | MGC21381 | transcription factor AP-2 beta (activating enhancer binding protein 2 beta) | - | HPRD | 7729426 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | - | HPRD,BioGRID | 12446731 |
| TP73 | P73 | tumor protein p73 | - | HPRD,BioGRID | 12080043 |
| TRRAP | FLJ10671 | PAF350/400 | PAF400 | STAF40 | TR-AP | Tra1 | transformation/transcription domain-associated protein | - | HPRD,BioGRID | 9708738 |
| TRRAP | FLJ10671 | PAF350/400 | PAF400 | STAF40 | TR-AP | Tra1 | transformation/transcription domain-associated protein | c-Myc interacts with TRRAP. | BIND | 12660246 |
| TUBA1A | B-ALPHA-1 | FLJ25113 | LIS3 | TUBA3 | tubulin, alpha 1a | - | HPRD | 7651436 |
| TUBA1B | K-ALPHA-1 | tubulin, alpha 1b | - | HPRD | 7651436 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 7651436 |
| TUBA4A | FLJ30169 | H2-ALPHA | TUBA1 | tubulin, alpha 4a | - | HPRD | 7651436 |
| TUBA8 | TUBAL2 | tubulin, alpha 8 | - | HPRD | 7651436 |
| XRCC5 | FLJ39089 | KARP-1 | KARP1 | KU80 | KUB2 | Ku86 | NFIV | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | Ku80 interacts with C-Myc origin. | BIND | 15910003 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | - | HPRD,BioGRID | 8266081 |
| ZBTB17 | MIZ-1 | ZNF151 | ZNF60 | pHZ-67 | zinc finger and BTB domain containing 17 | - | HPRD,BioGRID | 9312026 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER | 29 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDMAC PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2RB PATHWAY | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GLEEVEC PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VIP PATHWAY | 29 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARDIACEGF PATHWAY | 18 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TEL PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARF PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID CD40 PATHWAY | 31 | 26 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID MYC PATHWAY | 25 | 22 | All SZGR 2.0 genes in this pathway |
| PID IL2 PI3K PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID IL2 STAT5 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | 65 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA EGF SIGNALING UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR DN | 64 | 39 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA REVERSIBLY DN | 29 | 21 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER UP | 27 | 18 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| TAKADA GASTRIC CANCER COPY NUMBER UP | 7 | 5 | All SZGR 2.0 genes in this pathway |
| NUNODA RESPONSE TO DASATINIB IMATINIB UP | 29 | 20 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN | 45 | 34 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK1 AND MAPK2 TARGETS | 12 | 10 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK7 TARGETS | 8 | 8 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN RESPONSE TO MP470 DN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| BAKER HEMATOPOESIS STAT1 TARGETS | 10 | 9 | All SZGR 2.0 genes in this pathway |
| BAKER HEMATOPOIESIS STAT3 TARGETS | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BAKER HEMATOPOESIS STAT5 TARGETS | 7 | 7 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS AND SERUM RESPONSE DN | 47 | 34 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS AND SERUM RESPONSE UP | 47 | 32 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GENOTOXIC SIGNATURE | 105 | 68 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 SIGNALING 2 | 12 | 8 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 TARGETS WITH STAT5 BINDING SITES T1 | 9 | 7 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| MANN RESPONSE TO AMIFOSTINE UP | 20 | 12 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE NORMAL DN | 33 | 27 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE IMMORTALIZED DN | 31 | 26 | All SZGR 2.0 genes in this pathway |
| CEBALLOS TARGETS OF TP53 AND MYC UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS UP | 201 | 127 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| MANTOVANI NFKB TARGETS UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| SCIAN CELL CYCLE TARGETS OF TP53 AND TP73 DN | 22 | 14 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC UP | 33 | 20 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 2 | 30 | 23 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| TESAR ALK TARGETS HUMAN ES 5D DN | 7 | 5 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| SCHUHMACHER MYC TARGETS UP | 80 | 57 | All SZGR 2.0 genes in this pathway |
| DORSAM HOXA9 TARGETS DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA UP | 50 | 35 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| RORIE TARGETS OF EWSR1 FLI1 FUSION UP | 30 | 22 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS DN | 30 | 21 | All SZGR 2.0 genes in this pathway |
| JAIN NFKB SIGNALING | 75 | 44 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN AND NSC682994 DN | 15 | 12 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8 | 72 | 56 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| ZHENG RESPONSE TO ARSENITE DN | 18 | 15 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY UP | 45 | 29 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 3 UP | 18 | 11 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR DN | 47 | 31 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| MOREIRA RESPONSE TO TSA UP | 28 | 23 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 2HR | 51 | 36 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 1 | 33 | 24 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| VARELA ZMPSTE24 TARGETS UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C5 | 46 | 36 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| TRACEY RESISTANCE TO IFNA2 DN | 32 | 23 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| LEIN MEDULLA MARKERS | 81 | 48 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4 | 94 | 69 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 CORE DIRECT TARGETS | 19 | 14 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC | 58 | 43 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION DN | 105 | 67 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER UP | 73 | 53 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA | 51 | 35 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| CLIMENT BREAST CANCER COPY NUMBER UP | 23 | 20 | All SZGR 2.0 genes in this pathway |
| CONRAD STEM CELL | 39 | 27 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA DN | 100 | 63 | All SZGR 2.0 genes in this pathway |
| TAYLOR METHYLATED IN ACUTE LYMPHOBLASTIC LEUKEMIA | 77 | 52 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D UP | 89 | 62 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL UP | 51 | 32 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER BRCA1 UP | 36 | 18 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 MULTIPLE MYELOMA PROGRAM | 36 | 25 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE | 81 | 66 | All SZGR 2.0 genes in this pathway |
| ZHAN EARLY DIFFERENTIATION GENES DN | 42 | 29 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| CHIARETTI T ALL RELAPSE PROGNOSIS | 19 | 15 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR DN | 33 | 25 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 24HR UP | 12 | 6 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 32HR UP | 11 | 7 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR UP | 14 | 9 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 8 | 49 | 36 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR DN | 49 | 33 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR DN | 114 | 69 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-34b | 138 | 144 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-496 | 281 | 287 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.